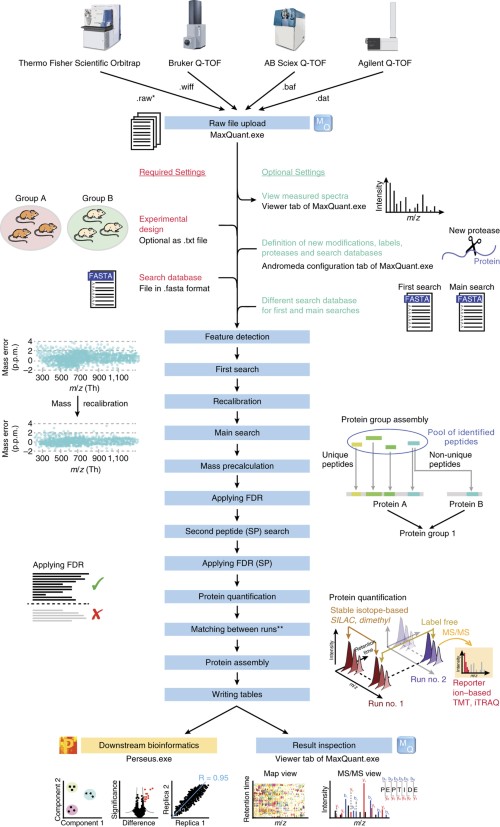
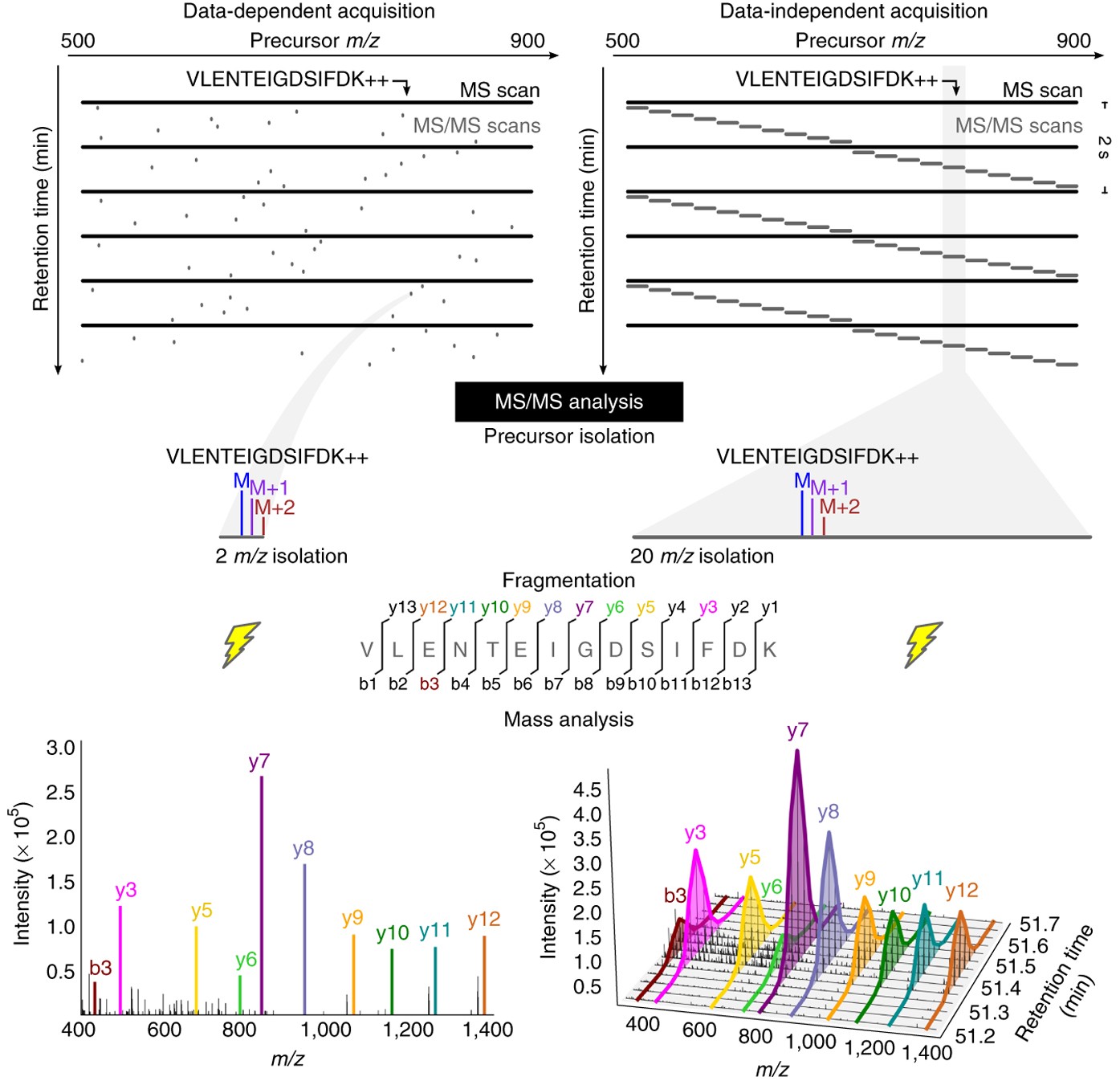
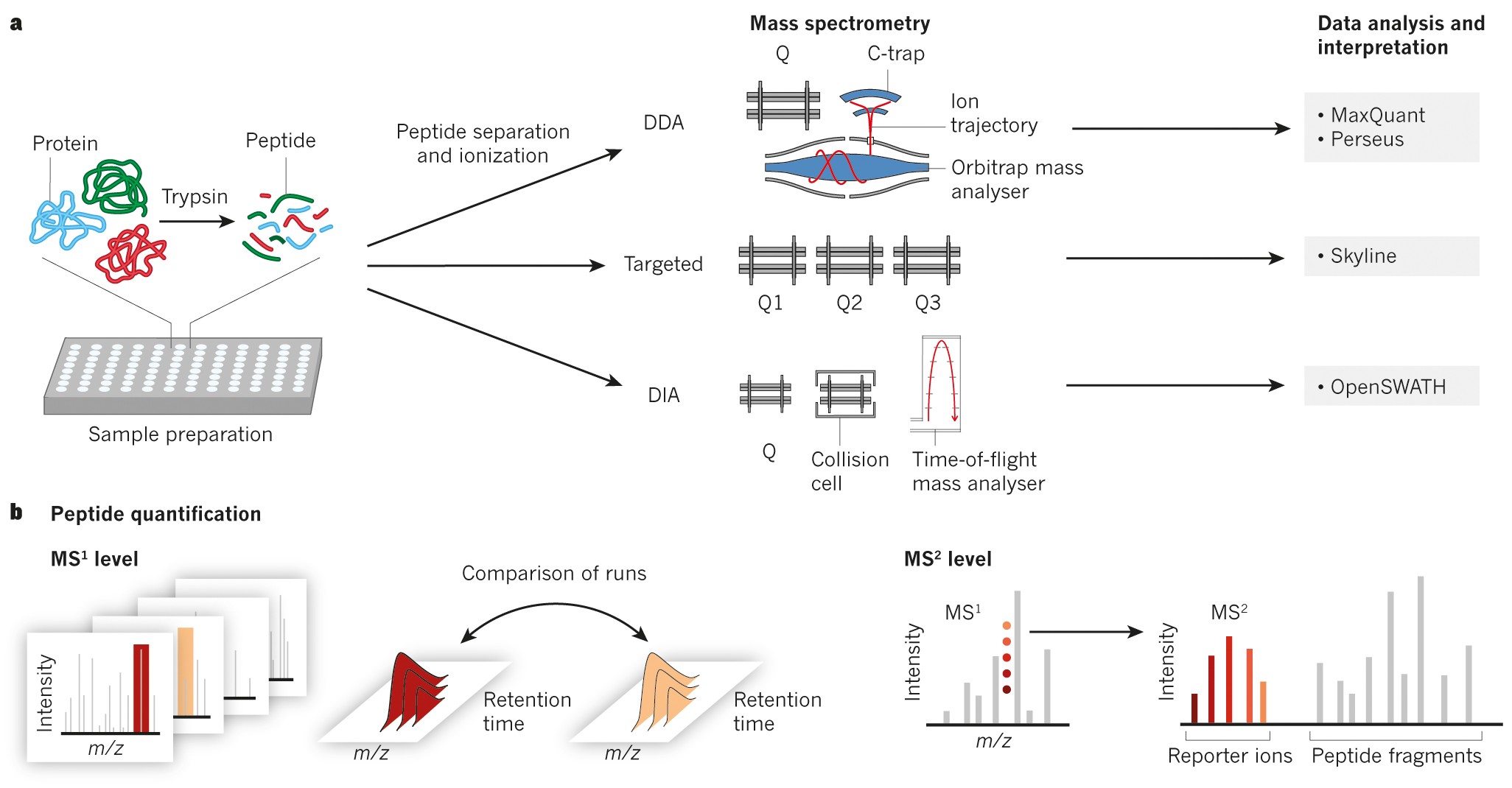
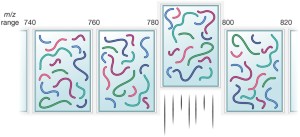
DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput | Nature Methods

Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics | Nature Communications

DIAproteomics: A Multifunctional Data Analysis Pipeline for Data-Independent Acquisition Proteomics and Peptidomics | Journal of Proteome Research

Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications
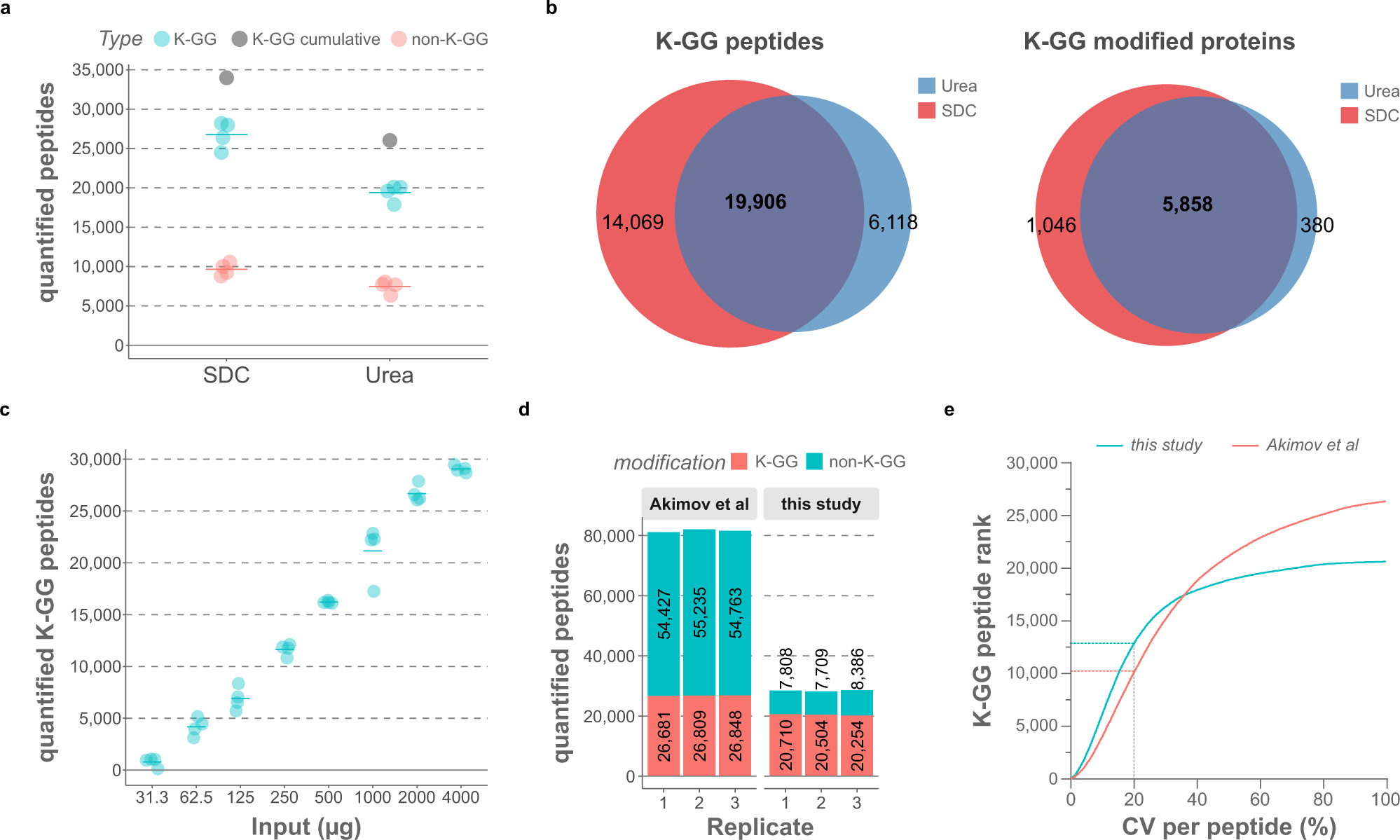
Time-resolved in vivo ubiquitinome profiling by DIA-MS reveals USP7 targets on a proteome-wide scale | Nature Communications
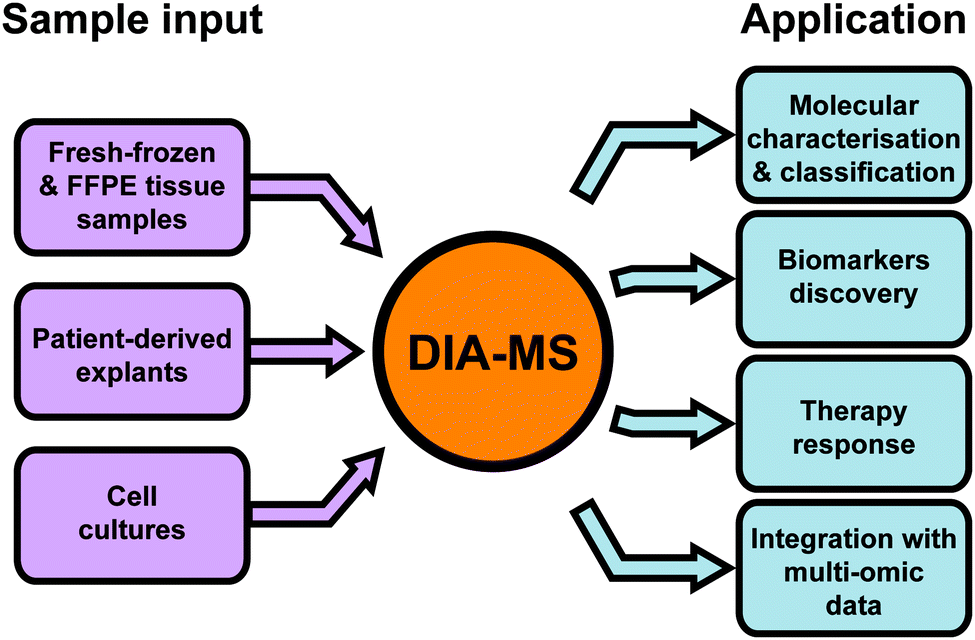
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H
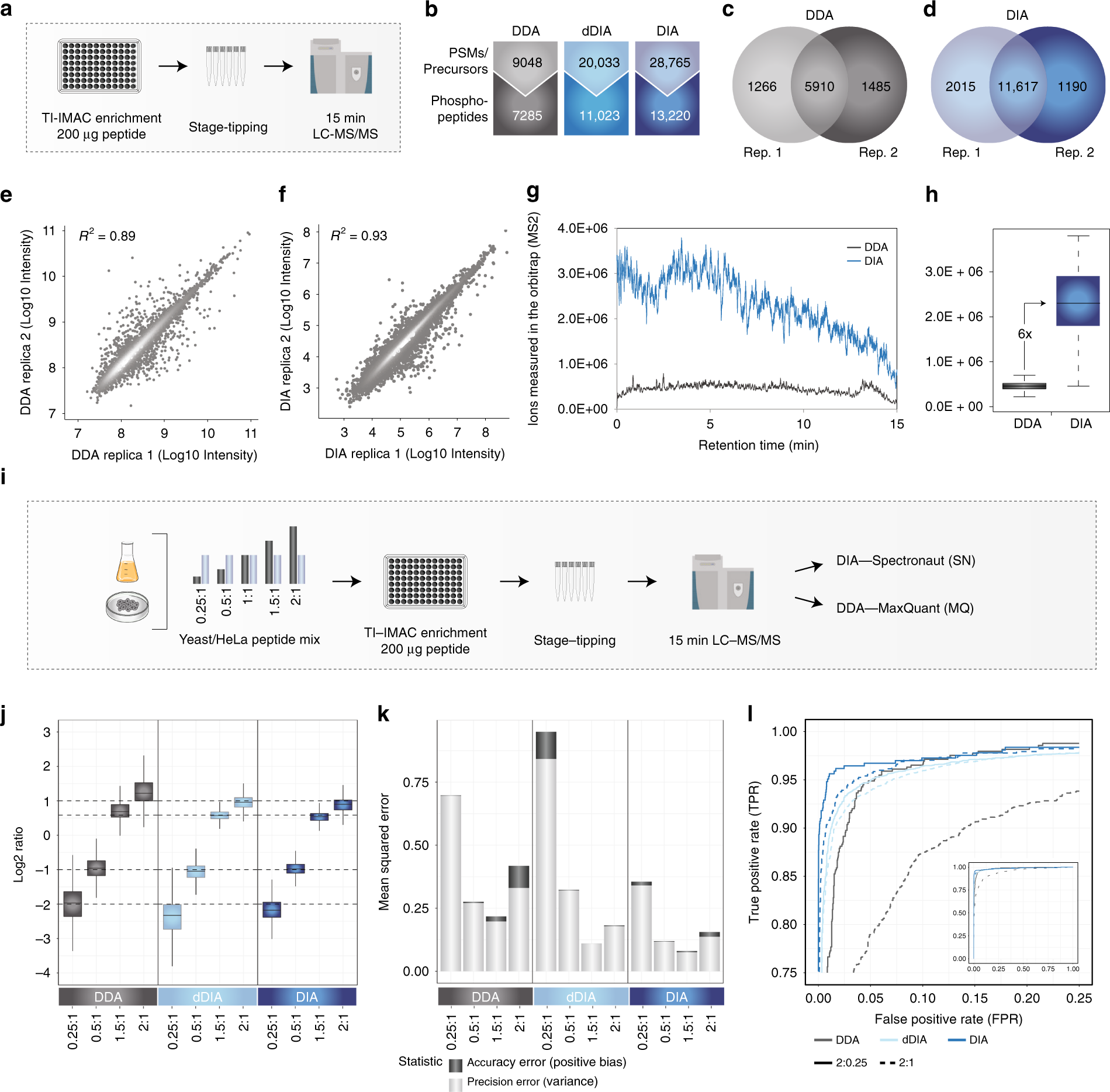
Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries | Nature Communications
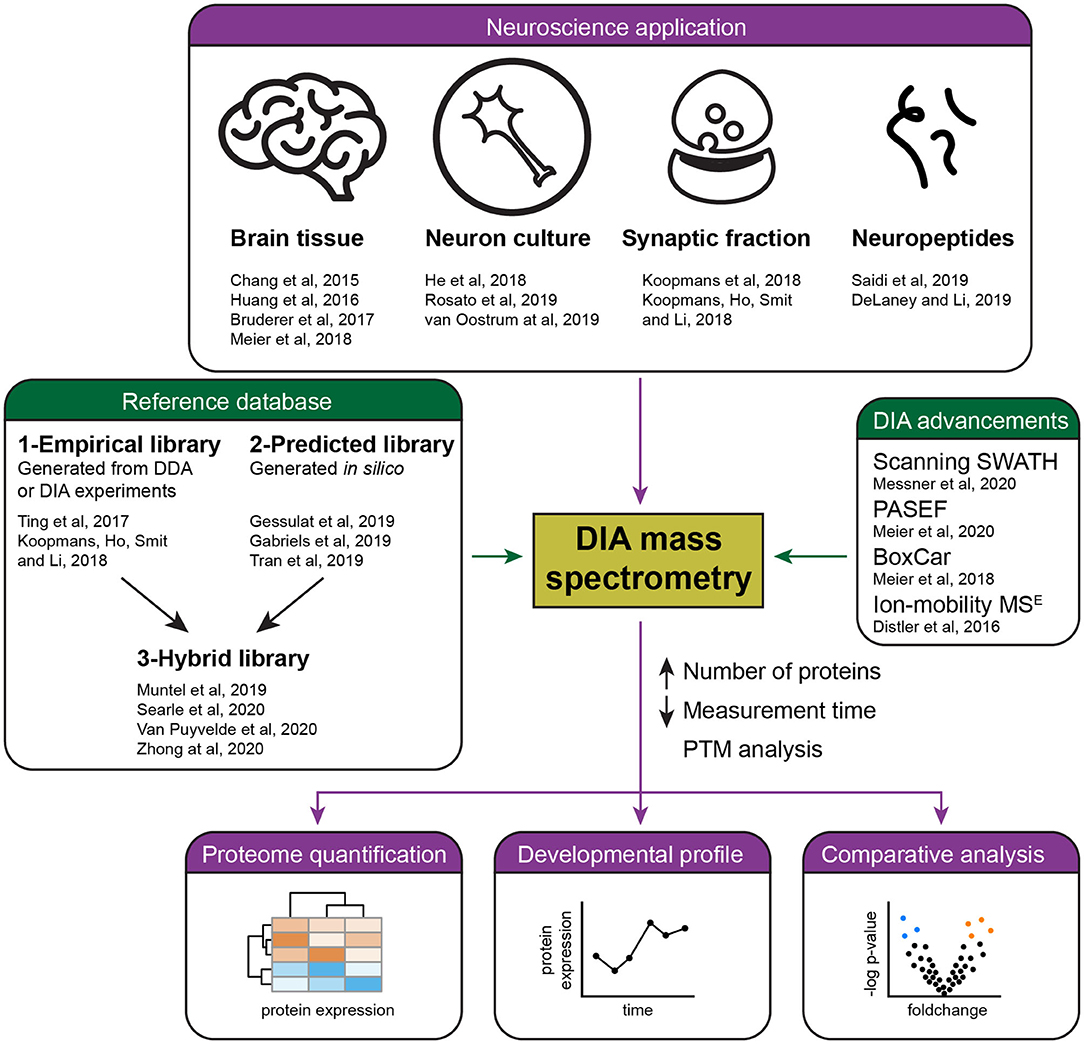
Frontiers | Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome

Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology | Nature Communications

Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files | Nature Methods
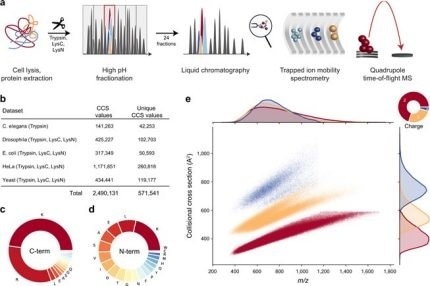
New Nature Communications publication by Mann & Theis Groups harnesses the benefits of large-scale peptide collisional cross section (CCS) measurements and deep learning for 4D-proteomics
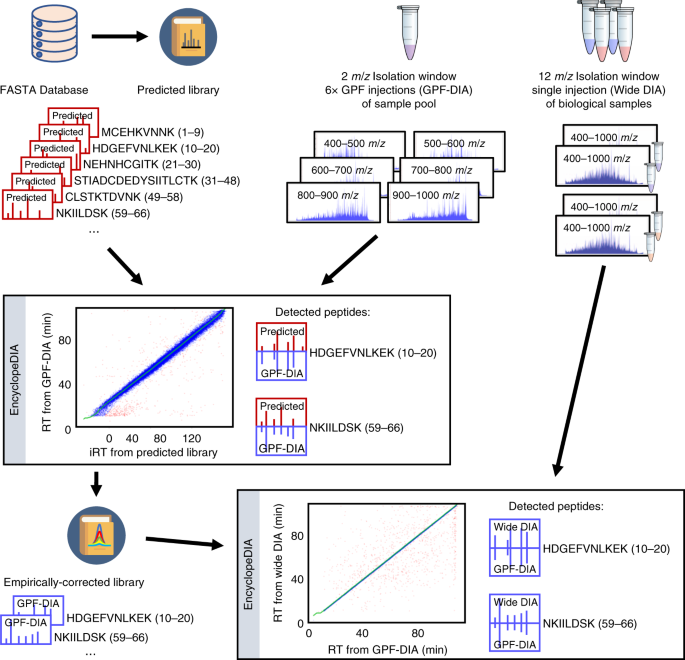
Generating high quality libraries for DIA MS with empirically corrected peptide predictions | Nature Communications

Deep Proteomics Using Two Dimensional Data Independent Acquisition Mass Spectrometry | Analytical Chemistry
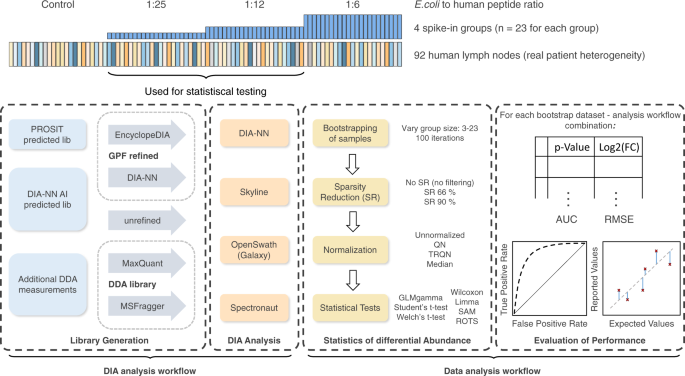
Benchmarking of analysis strategies for data-independent acquisition proteomics using a large-scale dataset comprising inter-patient heterogeneity | Nature Communications

Direct data-independent acquisition (direct DIA) enables substantially improved label-free quantitative proteomics in Arabidopsis | bioRxiv
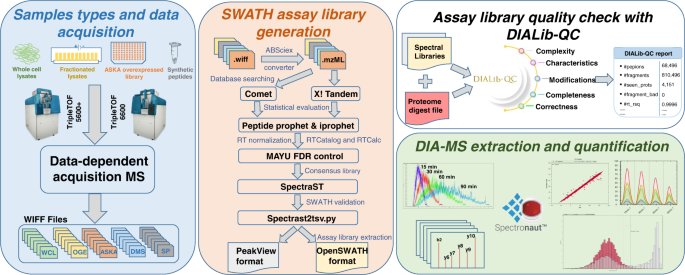
A comprehensive spectral assay library to quantify the Escherichia coli proteome by DIA/SWATH-MS | Scientific Data

Implementing the reuse of public DIA proteomics datasets: from the PRIDE database to Expression Atlas | Scientific Data
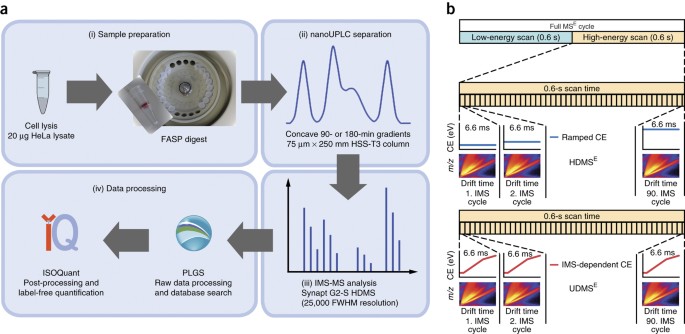
Label-free quantification in ion mobility–enhanced data-independent acquisition proteomics | Nature Protocols
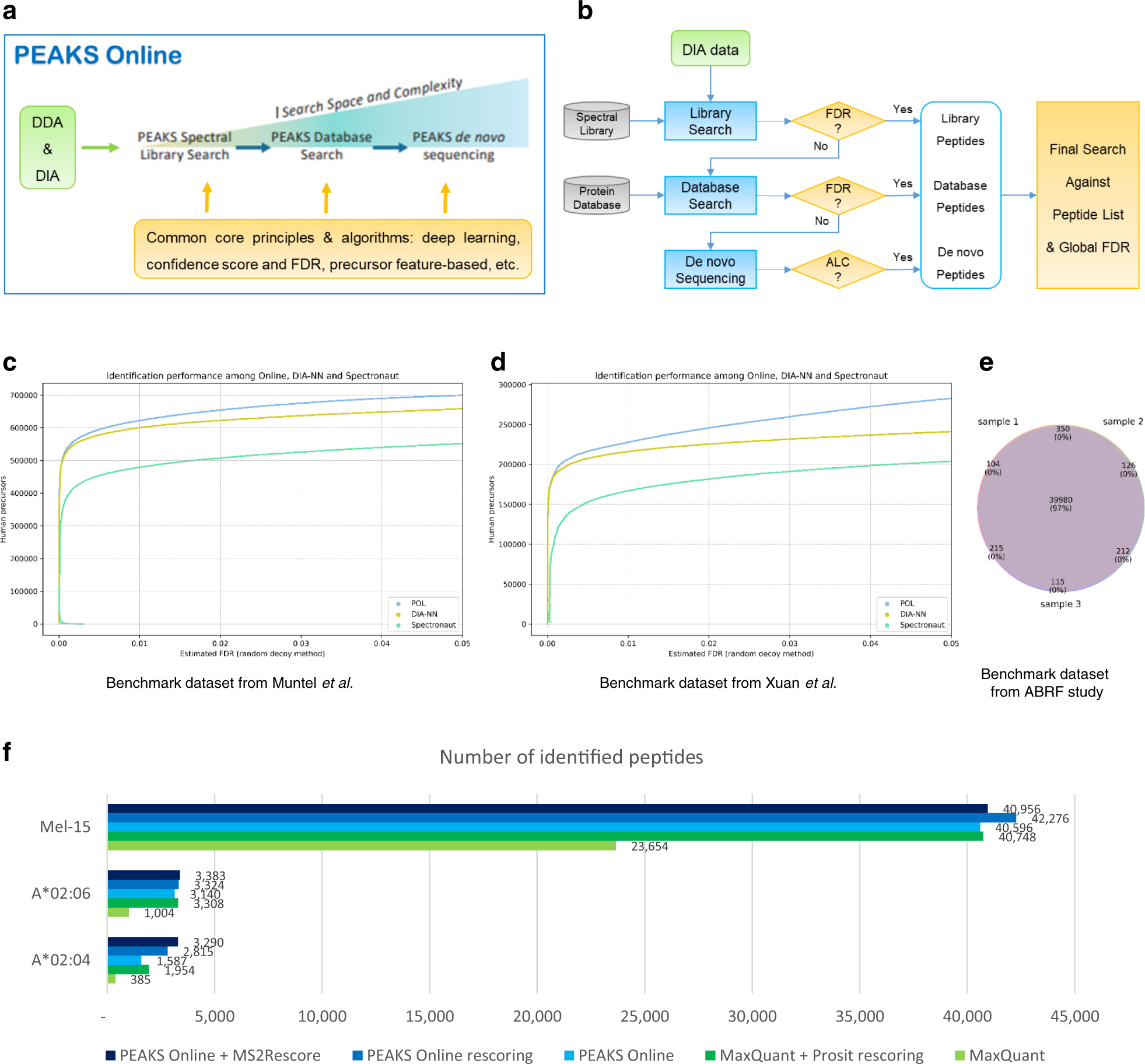
A streamlined platform for analyzing tera-scale DDA and DIA mass spectrometry data enables highly sensitive immunopeptidomics | Nature Communications
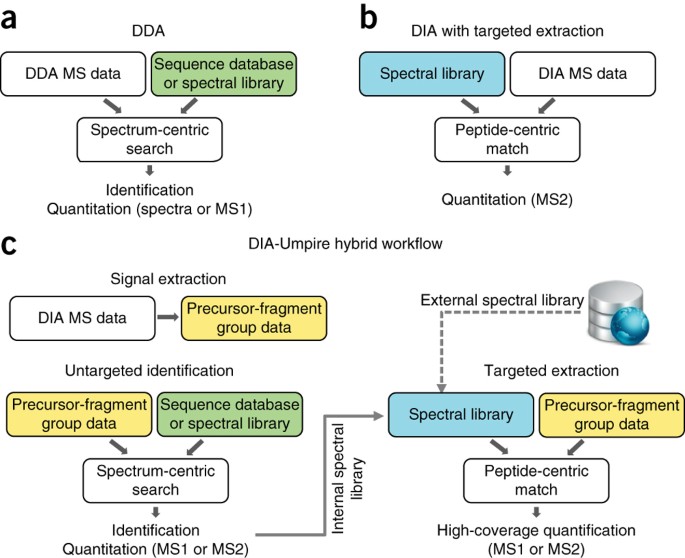
DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics | Nature Methods
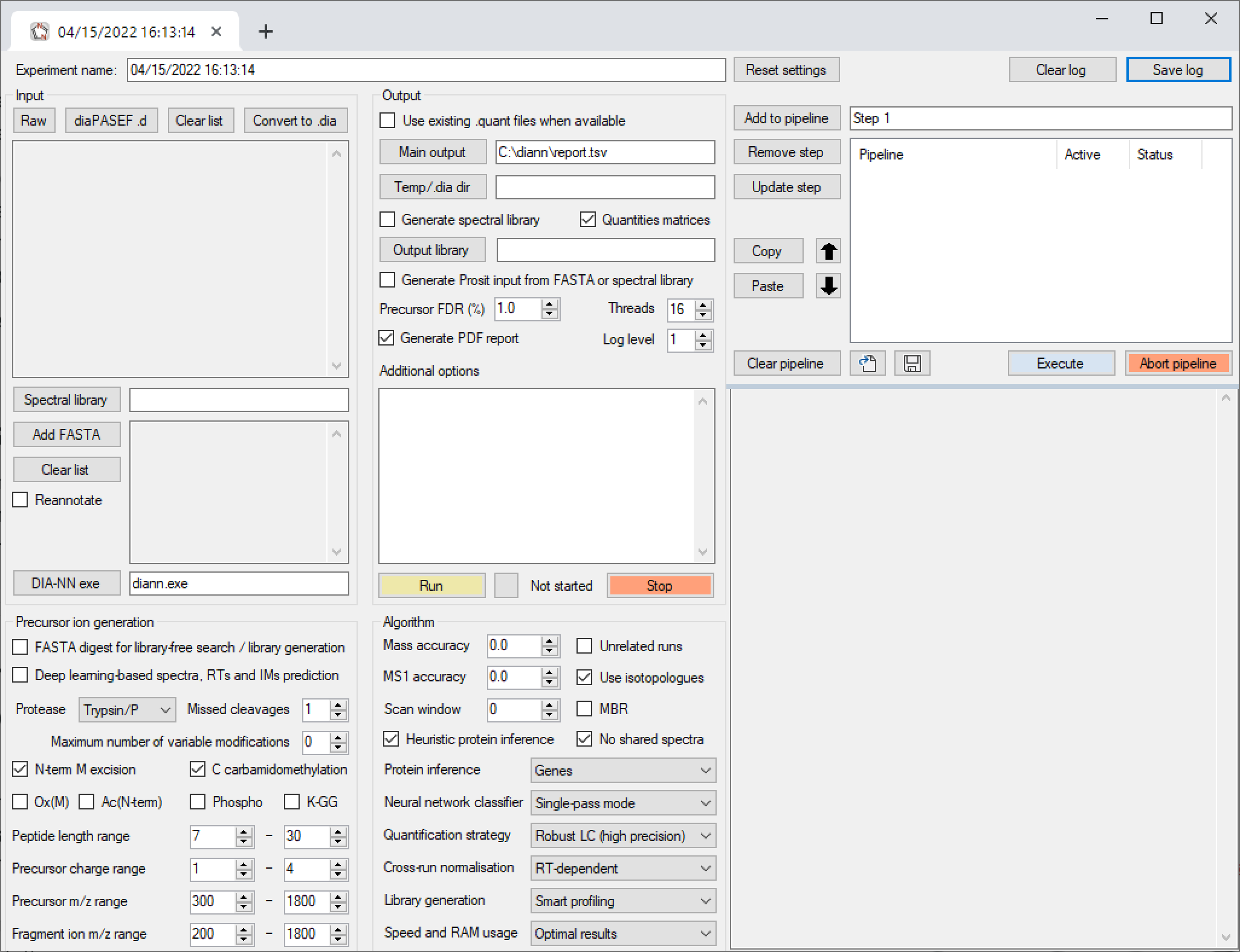
GitHub - vdemichev/DiaNN: DIA-NN - a universal automated software suite for DIA proteomics data analysis.